
Rediscovering Therapeutic Roots: How Ethnobotany Aids Drug Discovery
As our founder, Viswa Colluru, lays out in this blog post, the chemical code of life is equal parts powerful and mysterious. Our goal at Enveda is to crack this code by creating technologies that can do for life’s chemistry what Next Generation Sequencing did for genomics – and to use it to discover and develop the next generation of impactful nature-inspired medicines.
We’ve made significant progress towards building a platform that can determine the identity and function for every molecule within a sample in a high-throughput and multiplexed manner, without relying on the separation of individual molecules for analysis, a major step change for the field. But this advance raises an important question: where should we first deploy this NGS-for-chemistry tool to look for new medicines, given that life today consists of millions of species?
This brings us to another core observation at Enveda, which is that the fundamental problem in drug discovery is that candidate medicines that work in the lab don’t work in people. This issue with translation led us to ask: is there an underutilized source of medicines that we know already work in people? And the answer is a resounding yes.
People have been sampling nature in search of medicine for millennia. Such practices have taken root in every distinct culture throughout history, pre-dating Otzi the caveman. While Western science discounts these “traditional medicines”, many modern pharmaceuticals can be traced directly back to such practices (with morphine, aspirin, quinine, metformin, and artemisinin being key examples). Despite this, the accepted wisdom of most academics and pharmaceutical companies remains that traditional medicines and their associated cultural wisdom are at best difficult to replicate and at worst no better than placebo.
At Enveda, we value the generations of empiricism and experimentation that is represented in traditional medicine. We pay heed to the longevity of these practices despite the absence of “gold-standard” evidence. We are careful to not misconstrue the absence of evidence as the evidence of absence. At the same time, we are aware that these datasets and practices are noisy. By applying advances in data science, we believe that these datasets can be analyzed like any other large dataset, allowing for rank-ordering of testable hypotheses with best signal-to-noise ratio.
Over the past 4 years (and counting) we have collected, integrated, and analyzed the world’s traditional uses of plant-based medicine (i.e. ethnobotany) into a massive Knowledge Graph, which serves three key purposes:
- To collect, harmonize, and render computable the wisdom of our various cultures and recapitulate their ability to identify efficacious plant-based medicines through traditional practices
- To identify promising natural sources for interrogation using the Enveda platform by resolving patterns of shared usages across distant cultures
- To generate high-confidence therapeutic hypotheses that map novel leads that we discover from these natural sources to specific pathologies via distinct molecular pathways
In this post, we will share, for the first time, how we created the largest database of ethnobotany research and what our analyses revealed.
How we built this
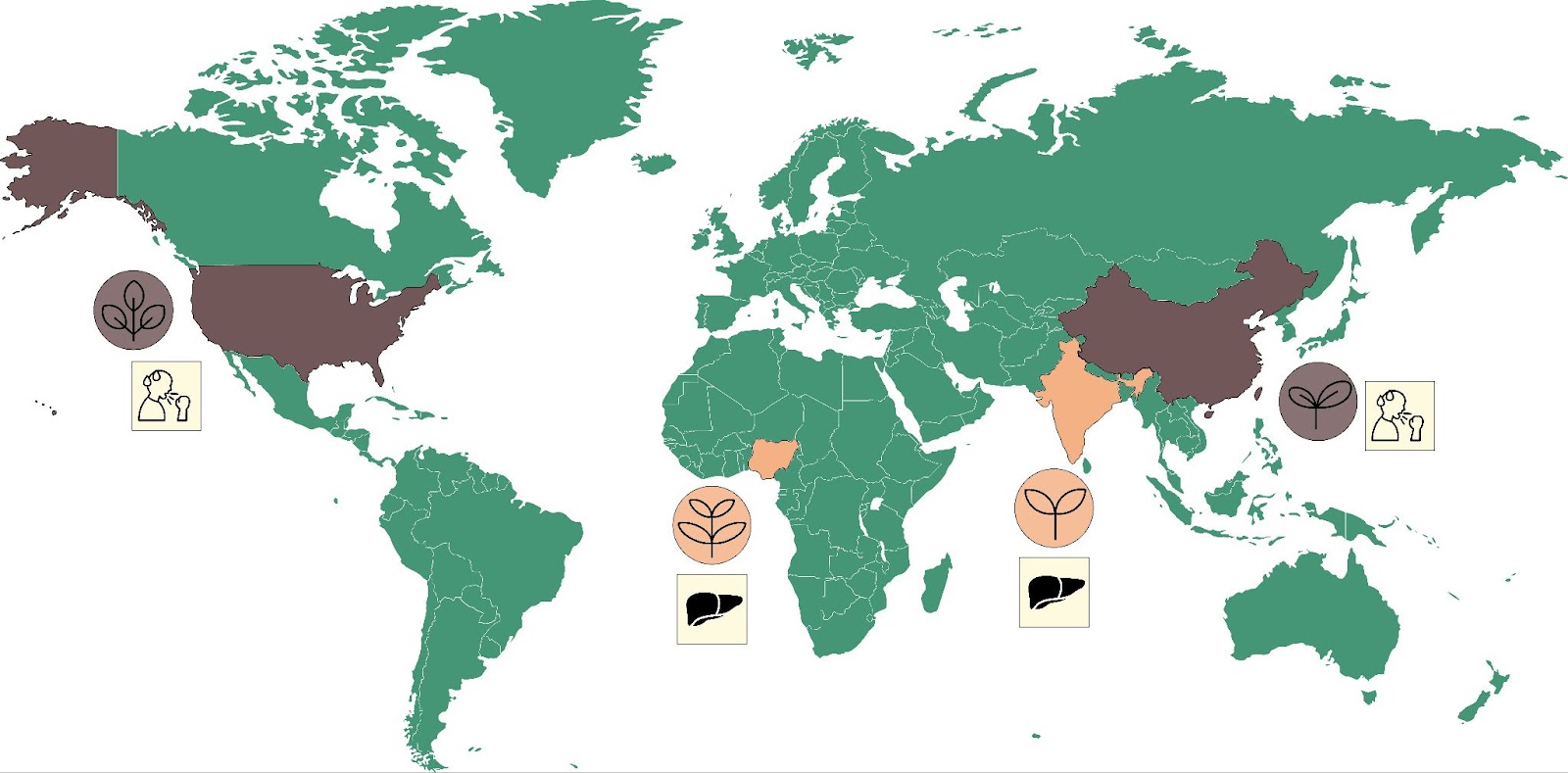
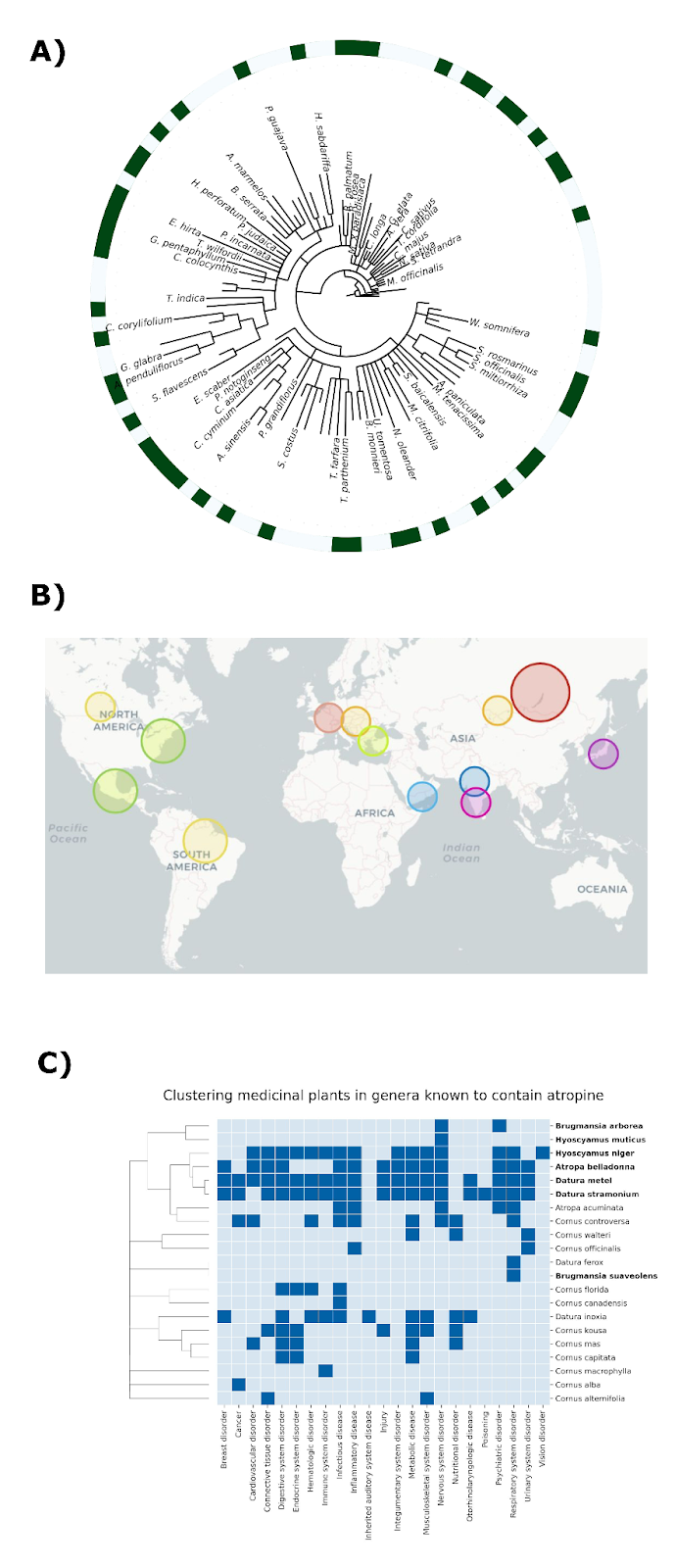
We are not the first team to study patterns of traditional plant uses across space and time. Previous work has identified anecdotal instances where related plant species are used to treat similar medical conditions despite growing in radically different environments on opposite sides of the world, as illustrated by Figure 1 [2].
Take for instance plants of the genus Tinospora. In India, Tinospora cordifolia is known as Guduchi and is used to treat a range of conditions including liver diseases and jaundice (see [3] for example). In Nigeria, Tinospora bakis is known as Aga Oyi and is also used to treat liver diseases and jaundice [4]. These are marked in Figure 1 in orange, and in brown is the example of China-native Gan Cao (Glycyrrhiza uralensis) and American licorice (Glycyrrhiza lepidota), which are both used to treat cough and sore throat [5].
While these types of anecdotes are illustrative, we wanted to undertake a more systematic study of these ethnobotanical patterns. As we describe in our recent publication [6], we leveraged a large language model similar to ChatGPT, paired with expert scientists, to systematically mine over 33 million scientific and ethnobotanical documents and extract thousands of associations between medicinal plants across all regions of the globe and therapeutic indications. Overall, the dataset captures over 5,000 plants from 140 countries in documents written over the course of five millennia.
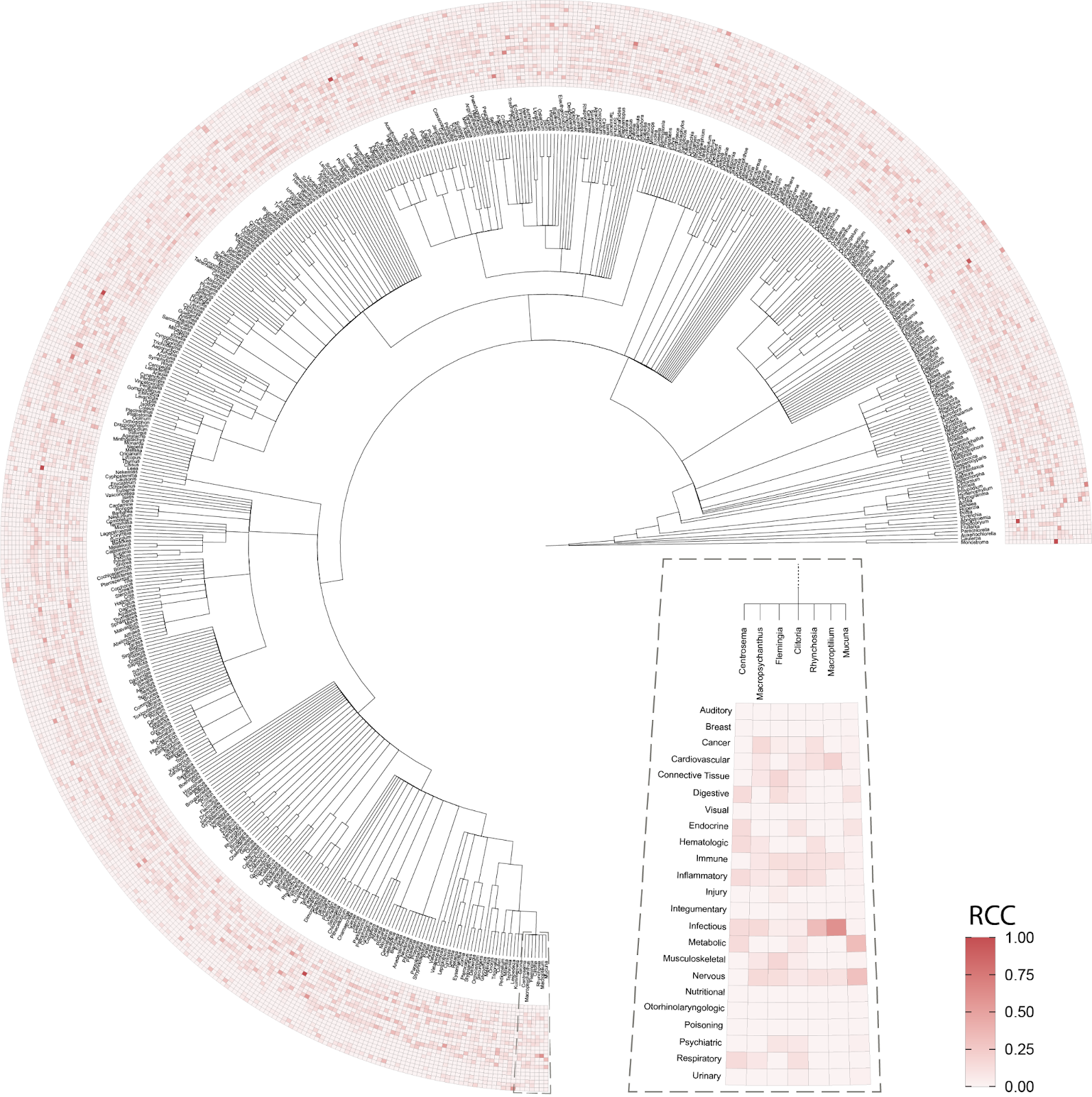
We then combined this database of ethnobotanical knowledge with two of the largest publicly available natural product databases representing the known chemical space of the plant kingdom (i.e., LOTUS [7] and COCONUT [8]) (Figure 2). This melding of ethnobotanical and chemical information allows us to investigate the potential bioactive compounds responsible for the therapeutic benefits of a given plant. However, we do note that most plant chemistry is unknown and so will not be represented in this database. To learn more about our efforts to explore the vastness of “dark” plant chemistry, please check out this blog post.
What we learned
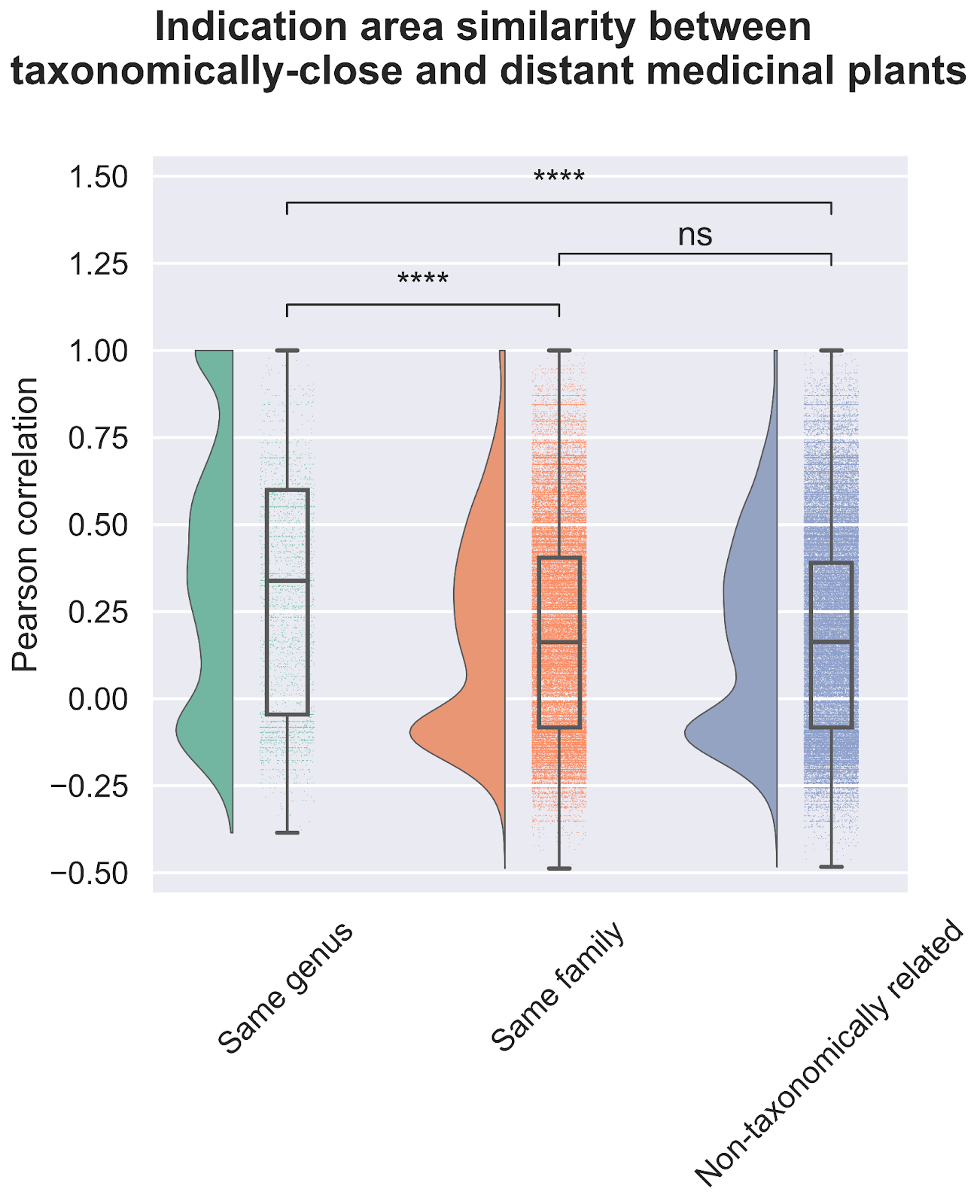
The first question we asked of our newly assembled knowledge graph was whether there was a random or non-random association between the use of related plant species for analogous medicinal purposes (Figure 3). A random association would suggest that traditional medicinal practices across geographic locations use species from throughout the taxonomic tree for therapeutic purposes, whereas a non-random association would suggest the potentially independent discovery of medicinal plants that are closely related within the taxonomic tree. Our results strongly point towards a non-random relationship. Medicinal plants within the same genus exhibited a statistically significant propensity to be used for treating similar health conditions when compared to plants from different genera; thus, supporting a biologically-rooted basis for the shared therapeutic properties of related plants.
Importantly, this strong relationship decreases when we look at the family level, suggesting a specific effect at the genus level. If you remember the Linnaean classification system from high-school, the taxa (or levels of relatedness) are ordered Kingdom, Phylum, Class, Order, Family, Genus, Species. When more lesser-related plants are added to the analysis by comparing at the Family level, we lose the similarity signal, again suggesting that related plants are used to treat related conditions.
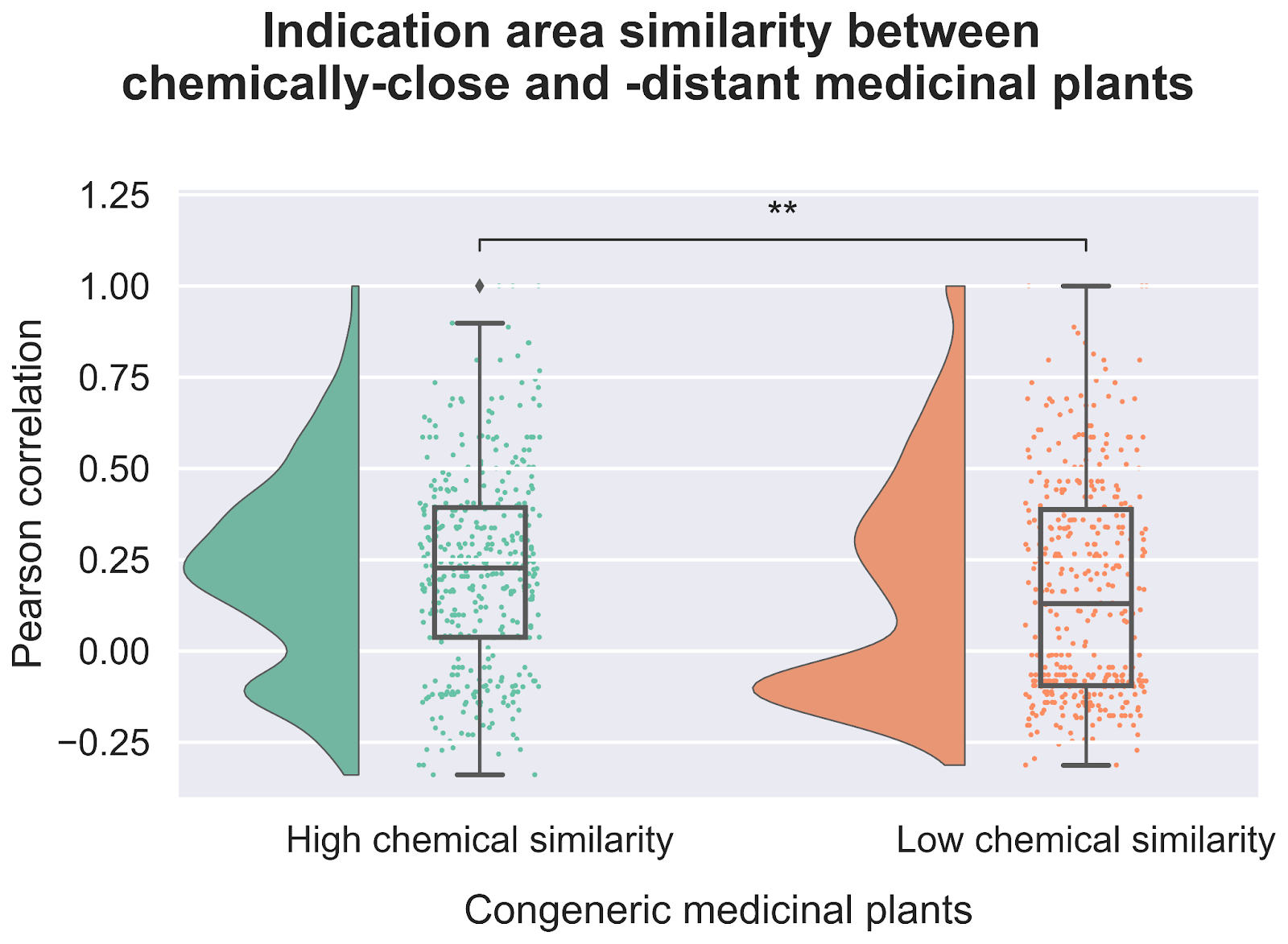
One possible explanation for our findings associating related medicinal plants to common ethnobotanical uses is that related species likely share conserved metabolic pathways and therefore produce either the same or similar metabolites. Metabolites are small molecules that are produced by living systems and that interact with the componentry of the cell, including proteins, RNA, and DNA. Metabolites are the main source of medicines from nature. Therefore, we explored the chemical space of these medicinal plants, hypothesizing that the shared therapeutic properties can be explained by the common presence of bioactive metabolites synthesized by their shared metabolic pathways.
Our exploration of the chemical space not only revealed that closely related plants tend to have similar chemical compositions, but also that there is a correlation between chemical similarity and therapeutic usage (Figure 4). When we looked at plants that were both evolutionarily related and had similar metabolite profiles we found an even stronger statistical relationship tying them to use treating similar diseases. This increases our confidence in the idea that common bioactive metabolites likely underlie the therapeutic effectiveness of these plants.
How we use this knowledge
As we mentioned at the start of this post, one of our goals for this work is to generate high-confidence hypotheses based on strong human priors for drug discovery.
The first and most obvious way we do this is by focusing on a particular indication, like asthma, and investigating which medicinal plants are most closely associated with it in our dataset. We were able to quickly identify several hotspots of closely related plants, which led us to the potential bioactive phytochemicals for this indication (Figure 5A). The most prominent hotspot was the Salvia genus, in which multiple species were strongly associated with asthma (i.e., reported in more than 15 publications). By clustering the species in this genus based on the chemical similarity of their metabolites, we were then able to pinpoint exactly which metabolites were exclusive to the species with the strong association to asthma. These shared metabolites, which include salvianolic acids, borneol, carnosol, and their many previously undiscovered metabolite analogs, are thus strong candidates for isolation and further screening in in vitro and in vivo assays.
A second, complementary, approach is to focus on the bioactive compound instead of the indication. Starting from a known bioactive chemical structure, we can use the combination of ethnobotanical and phytochemical knowledge to predict which diseases it is likely to remediate.
In our publication, we illustrate the utility of this approach with several examples. For instance, the sennosides are a group of chemicals often used to help with constipation and to clean the digestive system prior to surgery. We found sennosides in 6 different genera, namely: Cassia, Fallopia, Reynoutria, Rheum, Senna, and Terminalia. These 6 genera contain over 400 species, with 22 of them reporting to have therapeutic effects. Investigating the ethnobotanical knowledge around these 22 plants, we found that 8 of these plants contained sennosides, corresponding to the vast majority of the ethnomedicinal literature around digestive disorders, while the remaining plants that did not contain sennosides were weakly associated with these indications. Additionally, we found similar patterns for plants containing vinca alkaloids, a class of bioactive compounds used to treat cancer, as well as atropine, which is used for different health problems like issues with the nervous system, heart, and eyes. Overall, these examples illustrate how, by leveraging ethnobotanical knowledge, we can nominate high-confidence hypotheses on therapeutic areas after identifying a bioactive chemical and can start testing these compounds in disease models.
One common counter-argument to the value of this work is that perhaps this information on medicinal plants was shared between cultures, rather than independently discovered. While we can’t rule out this possibility directly for every shared usage, we can use the geographic information included in our database to generate compelling hypotheses. Thus, we looked at dozens of pairs of plants from the same genus with high correlation based on therapeutic usage, despite being located in parts of the globe where the native cultures that used them could not have had a high-degree of interaction (e.g., South America and Australia). Among these pairs of plants, we found several examples of both plants having been traditionally used to treat the exact same conditions, and we could also identify shared chemicals among these plants that exhibited bioactivity and are potentially responsible for this beneficial effect. These commonalities in spite of communication barriers strengthen our conviction that these plants have therapeutic benefits that were discovered through traditional practices.
Conclusion
Our knowledge graph of ethnobotanical practices maps out which plants, containing which compounds, have been used to treat which diseases in people. For the first time, computational organization and analysis of this knowledge has yielded strong evidence of empirical validity of ancient cultural practices. We believe that using this collective wisdom distilled from thousands of years of human experience as a starting point for our drug discovery process will help us to cross the lab-to-clinic divide.
Of course, this analysis is not without its drawbacks. As mentioned above, the chemical space represented in available databases only covers an estimated 1% of natural molecules, so the therapeutic effects seen with particular plants may be attributed to a yet unknown molecule. As such, we have also built a sophisticated metabolomics-based chemistry search engine for the natural world, as linked above. Moreover, the associations within this graph are correlative, not causative.
However, as we have utilized this database as part of our knowledge graph internally – in combination with our platform that annotates the structure and function of phytochemicals – we find a strong relationship between traditional plant usage and performance in our phenotypic screens. We are inspired to continue harvesting this resource for the next generation of nature-inspired medicines.
References
[1] Sun, D., Gao, W., Hu, H., & Zhou, S. (2022). Why 90% of clinical drug development fails and how to improve it?. Acta Pharmaceutica Sinica B, 12(7), 3049-3062.
[2] Moerman, D. E. (1991). The medicinal flora of native North America: an analysis. Journal of ethnopharmacology, 31(1), 1-42.
[3] Khare, C. P. (2008). Indian medicinal plants: an illustrated dictionary. Springer Science & Business Media.
[4] Pharmacopoeia, W. A. H. (2013). West African Health Organisation (WAHO). Bobo-Dioulasso (Burkina Faso).
[5] Quattrocchi, Umberto. CRC world dictionary of medicinal and poisonous plants: common names, scientific names, eponyms, synonyms, and etymology (5 Volume Set). CRC press, 2012.
[6] Domingo-Fernandez, D., Gadiya, Y., Mubeen, S., Bollerman, T.J., Healy, M.D., Chanana, S., Gura, R., Healey, D., Colluru, V. (2023). Modern drug discovery using ethnobotany: A large-scale cross-cultural analysis of traditional medicine reveals common therapeutic uses. iScience, Volume 26, Issue 9, 107729.
[7] Rutz, A., Sorokina, M., Galgonek, J., Mietchen, D., Willighagen, E., Gaudry, A., ... & Allard, P. M. (2022). The LOTUS initiative for open knowledge management in natural products research. Elife, 11, e70780.
[8] Sorokina, M., Merseburger, P., Rajan, K., Yirik, M. A., & Steinbeck, C. (2021). COCONUT online: collection of open natural products database. Journal of Cheminformatics, 13(1), 1-13.

.svg)

.svg)




